May 27 2010
Boston University researchers have developed a new technique that utilizes solid-state nanopores that could help in faster and cheaper DNA sequencing and thereby develop its use for routine clinical diagnostic applications.
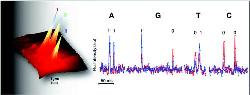 Boston University research on DNA sequencing
Boston University research on DNA sequencing
These nanopores are miniaturized holes in silicon chips for identifying the DNA molecules when they travel through the pore, resulting in recording the characteristics of the four nucleotides. These nucleotides encode every DNA molecule. The research team has also demonstrated the practicality of a new, more efficient technique that can identify individual DNA molecules in nanopores.
The Nano Letters at http://pubs.acs.org/doi/abs/10.1021/nl1012147 has provided a detailed report on the research.
This high-speed and low-cost DNA sequencing technique will be able to modernize biomedical and healthcare research, and could result in important improvements in personalized medicine, drug development and preventive medicine domain. A physician will be able to find out the possibility of a particular patient developing a genetic disease through access to the patient’s complete genome sequence.
The team has demonstrated through its findings that nanopores capable of examining very long DNA molecules very precisely are distinctively placed for competing with third-generation existing models for DNA sequencing techniques for accuracy, speed and cost. This new nanopore technique does not depend on enzymes unlike the existing approaches. The activities of such enzymes restrict the DNA sequences reading rates.
An application for a four-year grant is under consideration by the National Institutes of Health for further advancement of the nanopore sequencing project of Amit Meller, biomedical engineer from Boston University. Amit Meller has collaborated with other Boston University researchers and Worcester-based University of Massachusetts Medical School.
According to Meller, the cheap DNA sequencing technique could be introduced in the market in three to five years, by the adoption of a development program and aggressive research.
He revealed that the research team had utilized the optically enabled technique for DNA sequence readout along with the nanopore system for the first time. He clarified that the technique permitted the researchers to simultaneously probe multiple pores by utilizing just one speedy digital camera. He added that this enables the technique to scale up to a vast extent helping them in obtaining a high DNA sequencing throughput.
Source: http://www.bu.edu