Oct 4 2017
Designer biosensors capable of detecting antibiotic molecules of interest have been engineered by Researchers from North Carolina State University. The biosensors are considered to be a first step toward developing antibiotic-producing “factories” within microbes such as E. coli.
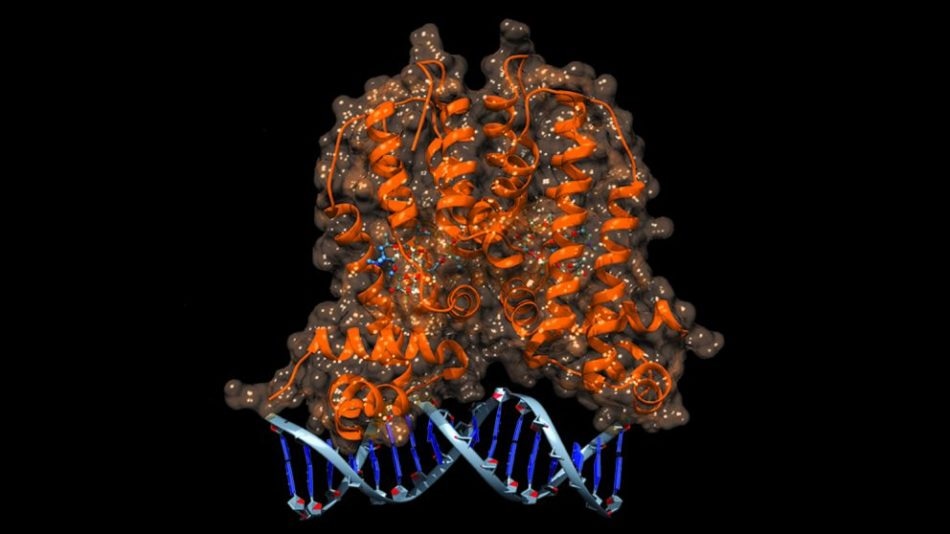 Illustration of the MphR biosensor binding to its target DNA sequence. Credit: Edward Kalkreuter
Illustration of the MphR biosensor binding to its target DNA sequence. Credit: Edward Kalkreuter
Macrolides are known to be a group of naturally occurring tiny molecules that can have antibiotic, anticancer or antifungal effects. The antibiotic erythromycin is one example – it is a macrolide developed by soil-dwelling bacteria. Researchers are interested in employing these natural antibiotics and the microbes that create them in order to produce new antibiotics; however, microbes that produce antibiotic macrolides only produce tiny amounts of a limited range of antibiotics.
Our ultimate goal is to engineer microbes to make new versions of these antibiotics for our use, which will drastically reduce the amount of time and money necessary for new drug testing and development. In order to do that, we first need to be able to detect the antibiotic molecules of interest produced by the microbes.
Gavin Williams, Associate Professor of Bio-organic Chemistry, NC State and Corresponding Author of a paper
A naturally occurring molecular switch – a protein known as MphR – was used by Williams and his team as their biosensor. In E. coli, MphR is capable of detecting the presence of macrolide antibiotics being secreted by microbes that are attacking E. coli. When MphR senses the antibiotic, it activates a resistance mechanism in order to negate the antibiotic’s effects.
The Researchers developed a large library of MphR protein variants and screened them for the potential to switch on production of a fluorescent green protein when they were in the presence of an anticipated macrolide. The variants were tested against erythromycin, which MphR already recognizes, and discovered that some of the MphR variants enhanced their detection ability tenfold. They also succeeded in testing the variants against macrolides that were not closely related to erythromycin, such as tylosin.
Essentially we have co-opted and evolved the MphR sensor system, increasing its sensitivity in recognizing the molecules that we’re interested in. We know that we can tailor this biosensor and that it will detect the molecules we’re interested in, which will enable us to screen millions of different strains quickly. This is the first step toward high-throughput engineering of antibiotics, where we create vast libraries of genetically modified strains and variants of microbes in order to find the few strains and variants that produce the desired molecule in the desired yield.
Gavin Williams, Associate Professor of Bio-organic Chemistry, NC State and Corresponding Author of a paper
The research has been published in ACS Synthetic Biology. The National Institutes of Health (grant GM104258) and the NC State Chancellor’s Innovation Fund funded the research. Graduate Student Yiwei Li, Former Graduate Student Christian Kasey, Former Undergraduate Student Mounir Zerrad, and T. Ashton Cropp, Professor of Chemistry at Virginia Commonwealth University, contributed to the work.